Reading SEER fixed width files
I help teach a class each summer which serves as an introducion to biomedical informatics and programming for, mostly resident, physicians. During the 4 weeks, we present basics in R and guide the students to start an analysis usually using a publically available dataset. One dataset that comes up every year is SEER from the National Cancer Institute.
But, there’s a problem. SEER suggests you use their software, SEER*Stat, which requires you to use Windows. They do provide data in an alternative “ASCII text format” but provide little guidance for new users to get started.
Reading text files
Reading data into R is full of pitfalls for any new programmer:
- multi-line headers,
- atypical delimiters,
- odd field types (e.g. Excel dates).
- leading zeros / whitespace,
- atypical values for missing data,
- …
SEER throws another curveball because their “ASCII text format” is actually a fixed width file!
Fixed width files
Most text based data files use a delimiter (otherwise known as a separator; e.g. commas, semicolons, spaces, or tabs) that marks the end of one field and the start of the next. On the other hand, a fixed width file (FWF) specifies the position or width of each field.
For example, here is a file where the first field is 20 characters long, the second field is 10 characters long and the third, and final, field is another 12 characters long.
John Smith WA 418-Y11-4111
Mary Hartford CA 319-Z19-4341
Evan Nolan IL 219-532-c301
To make FWFs even more complicated, they often don’t use a header to specify the field names. Since the user is supposed to know the field positons, they should also know the field names.
With the field names and positions (or widths), we can load the file and it should look like this:
| Name | State | SSN |
|---|---|---|
| “John Smith " | “WA " | “418-Y11-4111” |
| “Mary Hartford " | “CA " | “319-Z19-4341” |
| “Evan Nolan " | “IL " | “219-532-c301” |
R code
The readr::read_fwf() function is great for reading FWFs.
The format is very similar to the rest of readr (and, in turn, the rest of the tidyverse) and you can specify the field names and positions in your favorite way or let read_fwf guess for you.
library(readr)
read_fwf(fwf_sample, fwf_cols(name = 20, state = 10, ssn = 12) )
Which returns a tibble that looks like:
# A tibble: 3 x 3
name state ssn
<chr> <chr> <chr>
1 John Smith WA 418-Y11-4111
2 Mary Hartford CA 319-Z19-4341
3 Evan Nolan IL 219-532-c301
You’ll notice that read_fwf trimmed the whitespace from the end of each field too ("John Smith" rather than "John Smith "), which is nice.
Reading SEER data
So, how do you apply this to SEER if we don’t know the column names and positions?
You can’t.
But, each SEER “ASCII text file” comes with a .sas script that can be used to read the file into SAS.
Here’s an example that looks like this:
*filename seer21 './yr2000_2016.seer21/*.TXT';
data in;
infile seer21 lrecl=87;
input
@ 1 PUBCSNUM $char8. /* Patient ID */
@ 9 REG $char10. /* SEER registry */
@ 19 RACE1V $char2. /* Race/ethnicity */
@ 21 SEX $char1. /* Sex */
...
Now I don’t know SAS but I can see what this code is doing.
Each line after input defines one field and it’s:
- start position,
- field name,
- field type (and length/precision [the number after “char”]), and
- a bonus description of the field.
With this .sas file we have everything we need to read the entire file.
Programmatically extracting field information
We can use R to extract the 4 components that describe each field in the .sas file.
Using tidyverse again here:
sas.raw <- read_lines(sas.file)
sas.df <- tibble(raw = sas.raw) %>%
## remove first few rows by insisting an @ that defines the start index of that field
filter(str_detect(raw, "@")) %>%
## extract out the start, width and column name+description fields
mutate(start = str_replace(str_extract(raw, "@ [[:digit:]]{1,3}"), "@ ", ""),
width = str_replace(str_extract(raw, "\\$char[[:digit:]]{1,2}"), "\\$char", ""),
col_name = str_extract(raw, "[[:upper:]]+[[:upper:][:digit:][:punct:]]+"),
col_desc = str_trim(str_replace(str_replace(str_extract(raw, "\\/\\*.+\\*\\/"), "\\/\\*", ""), "\\*\\/", "" )) ) %>%
## coerce to integers
mutate_at(vars(start, width), funs(as.integer)) %>%
## calculate the end position
mutate(end = start + width - 1)
(I’m assuming sas.file is either a link to a file online, such as "https://seer.cancer.gov/manuals/read.seer.limitedfield.research.nov2018.sas", or a local file, such as "path/to/file/example.sas")
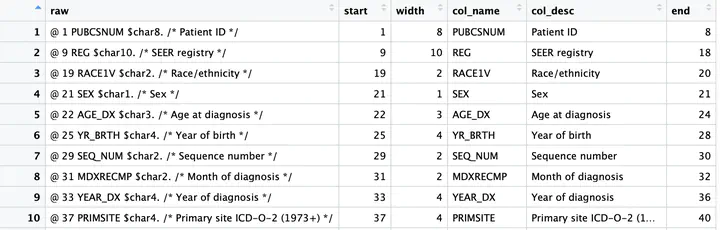
And with that we have a sas.df data frame (or tibble) that looks like:
# A tibble: 35 x 6
raw start width col_name col_desc end
<chr> <int> <int> <chr> <chr> <dbl>
1 " @ 1 PUBCSNUM $char8. /* Patient ID */" 1 8 PUBCSNUM Patient ID 8
2 " @ 9 REG $char10. /* SEER registry */" 9 10 REG SEER registry 18
3 " @ 19 RACE1V $char2. /* Race/ethnicity */" 19 2 RACE1V Race/ethnicity 20
4 " @ 21 SEX $char1. /* Sex */" 21 1 SEX Sex 21
Most importantly, sas.df contains start (and width) values and names for each field that we can directly use to read the whole file:
## read the file with the fixed width positions
data.df <- read_fwf(file_path,
fwf_positions(sas.df$start, sas.df$end, sas.df$col_name))
And we’re done!
Summary
I’ve used this template to help several different people start different SEER projects over the years and as long as you can find a .sas file that matches your data, this will work with almost no effort.
However, different SEER datasets have different formats so take care to check.
A simple script is up on github here.
Don’t forget to change the .sas file/link and the file path to your own copy of whichever SEER dataset.